Polygonal DNA origami
Since its inception in 2006, DNA origami has mainly relied on packing DNA double-helices side by side like in a log house. This made it easy to rout the long single-stranded scaffold and provides block-like structures that are in general quite rigid. We are working on ways to make the design process more free, so that a user can draw a polygon mesh in a 3D software and render these meshes directly in DNA – without the need to ‘blockify’ the design as well as providing a way to make . The scaffold routing turned out to be quite complex in these new structures, we solved that by employing mathematical tricks like making meshes Eulerian and then routing using A-trails. The result is a robust technology, first demonstrated in our paper in Nature in 2015, which has proven to have distinct advantages in physiological conditions compared to classical block-like origami, see for example our study in Advanced Materials from 2021.

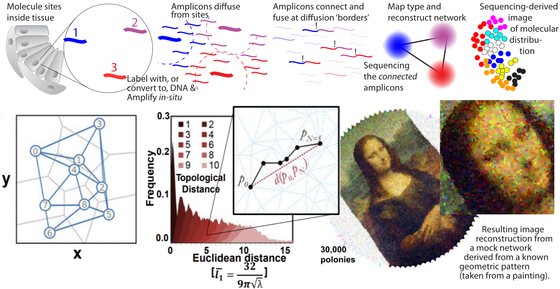
Imaging by sequencing
We are working on methods to reconstruct geometrical data of molecular locations based on sequencing alone. The state-of-the-art high throughput sequencing technologies can produce data on a massive scale – and the capabilities are ever increasing year by year. We hypothesize that in addition to information about DNA sequences, we can use these technologies to measure spatial data. If molecules are tagged with DNA (or just converted to DNA if they were RNA) we can drive reaction networks that produce neighbor-to-neighbor information encoded in the resulting DNA. When this DNA is sequenced, a computer can deduce the network information and reconstruct an image of the original molecular locations. Image shows a rough outline and results from our published work on DNA sequencing microscopy in PNAS from 2019.
DNA Nanocalipers for Cell Signaling Studies
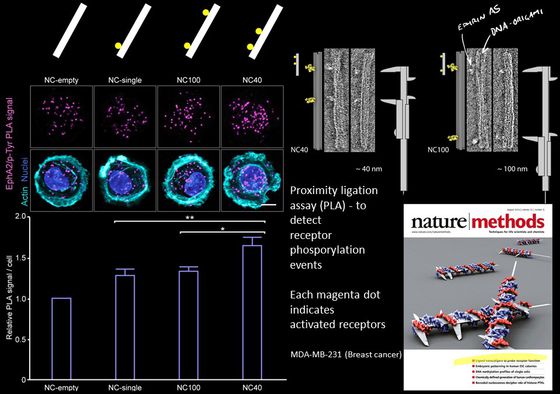
Mammalian cells signal to each other using proteins at their membrane surface. One hypothesis is that they can read patterns in neighboring cells and then react differently to different patterns of the same protein. You could think of it as the cells having a kind of tactile alphabet, like the Braille system for blind reading, but instead of recognizing dots on a paper. Cells read patterns of proteins in neighboring cells.
Actually testing this hypothesis and examining this “tactile alphabet” of cells has been very difficult because it was always very hard to precisely pattern proteins on the nanoscale and even harder to do it in suspension (i.e. not patterned on a surface). With DNA origami we can make very precise patterns of proteins ligands and study the effect the patterns have on receptor activation. In our published work we have been looking at the Ephrin/Eph pathway. Currently we are focusing on other receptor-ligand systems such as Notch.
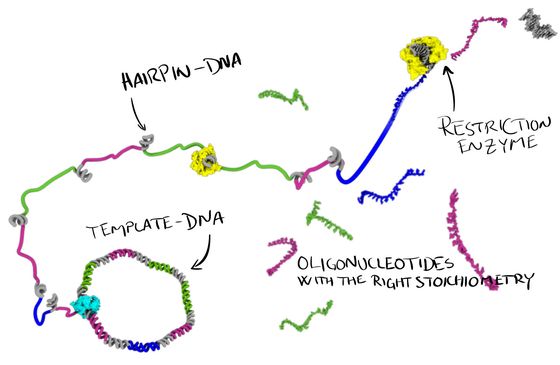
Enzymatic DNA oligonucleotide production
Our lab is working on new ways to produce single-stranded DNA oligonucleotides. In our ‘MOSIC’ method (Monoclonal Stoichiometric), we use a cloned DNA template to encode our oligos. From this template we produce single-stranded DNA, some of it programmed to fold up into hairpins that can be digested by a restriction enzyme and thereby releasing these very high quality, monoclonal, oligonucleotides in perfect stoichiometry.
The approach will be useful, not only for DNA nanotechnology, but also for anyone else that needs very pure, high quality DNA oligonucleotides for their research. The method can also produce very long oligos, we have demonstrated up to 400 nt, and longer should definitely be possible.
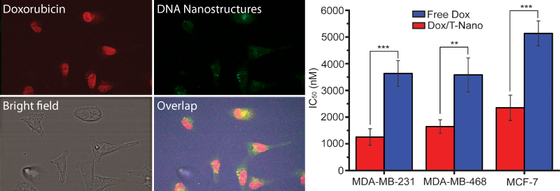
Using DNA Nanostructures for Drug Delivery
We are interested in using DNA nanostructures to deliver drugs. Previously we have been able to show that by using DNA origami, we are able to deliver the anti-cancer drug doxorubicin more efficiently than by using the free drug. In cell studies, the same effect could be achieved using less drug than when using the drug alone.
In the future we would like to explore if it is possible to combine this approach, with cancer-cell specific targeting and ways of protecting the DNA nanostructures from the immune system.
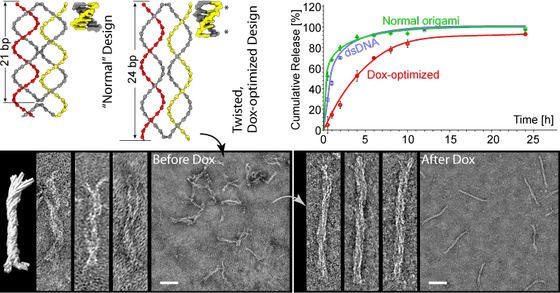
By designing the DNA nanostructures so that they force the DNA to twist and unwind, we are able to load more Doxorubicin (Dox) and control its release over a longer period.
Similar approaches could be tried with other drugs, and be used as a way to dock other types of small molecules. We are also interested in specific, nanoscale, patterning of proteins and other nanoparticles on the surface of our structures.
Björn presenting this work at the CLINAM conference. (Note, wrong title of the YouTube video).
Using DNA as a nanoscale printer.
DNA is a fantastic material. It’s information-rich, small in size and it glues itself together in a very predictive way. Using these three attributes, many researchers around the globe are now able to produce nanoscale structures that build themselves in test-tubes by simply mixing different types of DNA molecules.
Nanoscale construction using DNA has been around for a few years, Ned Seeman introduced the idea back in 1982. But it is only recently that a technique to routinely manufacture complex 3D shapes has evolved. Now you can design your desired 3D nanostructure on your computer, order the DNA, mix it with some magnesium salt, heat it up, cool it down and pour out your creation.
To render our structures in DNA we usually use a method called DNA origami, first reported by Paul Rothemund at Caltech in 2006 and further developed in William Shih’s lab and Kurt Gothelf’s lab in the final years of the last decade.
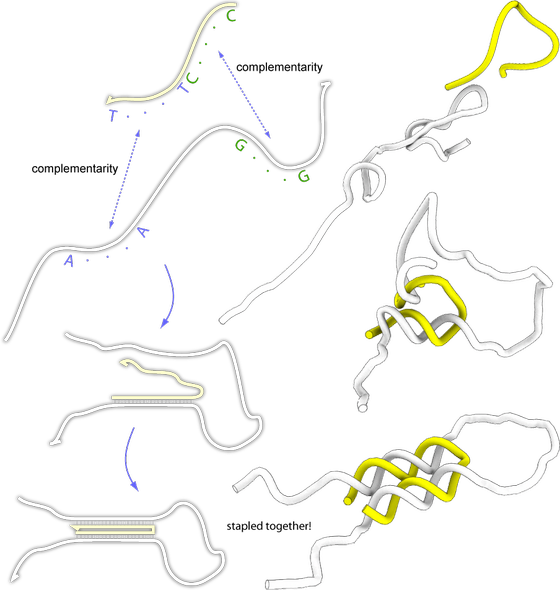
Lets say we would like to glue together two parts of a DNA molecule and the two parts are not complementary, can we do this? There is a trick we can use, lets design a smaller DNA molecule that have regions that are complementary to the two regions and use this to bring the two parts of the longer molecule together. Like this:
In this figure, the white strand is the one we would like to glue together. The yellow strand is the strand we designed so that it would bind the two regions of the white strand together. As you can se, it is possible to fold any DNA strand with the help of another strand.